This article is a summary of this video: “Nuclear receptor NR4A is required for patterning at the ends of the planarian anterior-posterior axis”. https://jrnlclub.org/research-films/planarian-regeneration-patterning
Regeneration is a fundamental process in biology that allows animals to “bounce back” from injury by synthesizing new tissue. While regeneration is an essential process for the maintenance of homeostasis in organisms, it is equally as important that regeneration is regulated and informed by the biological context that is situated within. Regenerative processes like cell proliferation that go awry contribute to cancer (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2706275/), and at a more basic level, if you lose an arm then you’d want an arm to grow back, not a leg or a head. While that seems like a silly question to think about concerning humans, this can definitely be an issue for organisms that are highly regenerative.
Regeneration can be more extensive in some organisms than others. Some mammals like mice (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7055364/) have some limited regenerative capabilities, but if you study organisms that are more evolutionarily distant from humans some animals like axolotls, hydra, and planarians have incredible regenerative capabilities.
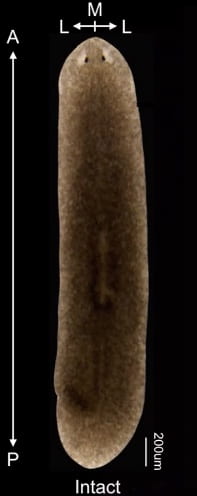
Planarians are flatworms that have what is called “dorsoventral polarity”. In other words, they have a clearly defined head and tail region: the head having two eyespots that detect light. At the molecular level, different characteristic genes are expressed at the head and tail regions.
They are famous for their regenerative capabilities because they are able to perform whole-body regeneration: (https://www.imrpress.com/journal/FBL/13/16/10.2741/3160) chop a planarian into 200 little pieces and each tiny piece can regrow tissue until it is its own little planarian, with all of the structures normally found in planarians like the head and tail.
Here is a video of a planarian regenerating its body after having its head amputated! (https://www.youtube.com/watch?v=hTC1eNTBXvE)
Scientists all over the world are interested in understanding how planarians regenerate the right tissue in the right places. Part of the answer is summarized in the phrase “positional information”: the presence of genes or groups of genes can delineate regions in the planarian body, or provide information about the part of the body that they are contained in. These genes are called positional control genes, or PCGs.
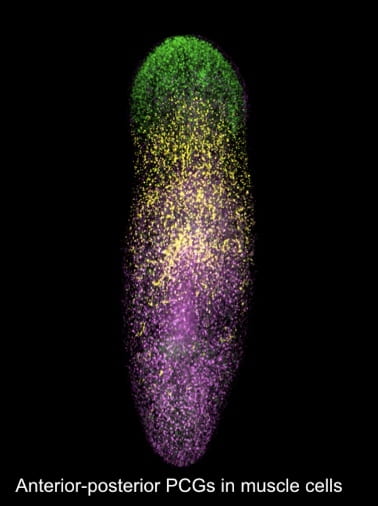
Dayan Li, working with Conor McMann and Dr. Reddien at the Whitehead institute, aimed to investigate the genetic markers that determine the way in which planarian regeneration occurs using imaging techniques and RNA sequencing.
As planarians are polarized along their dorsoventral axes, they have poles as well at the ends of their bodies: the tips of their heads and tails respectively. Li, McMann, and Reddien were interested in seeing if pole regions are “organizers” for head and tail identity. In other words, they wanted to see if the tissue found in these regions can direct the organized synthesis of new tissue. To answer this question, anterior pole tissue was transplanted into posterior regions of planarians and head-like outgrowths with eyes were generated, suggesting that anterior pole tissue induces surrounding tissue to become “head” tissue.
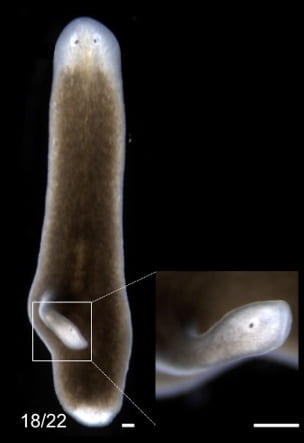
A whole concert of genes is present at the anterior and posterior poles, and the gene that was highlighted in this video was nr4A, or nuclear receptor 4A. Li, McMann, and Reddien showed that when RNA interference was used to disrupt the expression of nr4A, multiple “ectopic” pairs of eyes appear in planarian heads where wild-type planarians possess one pair.
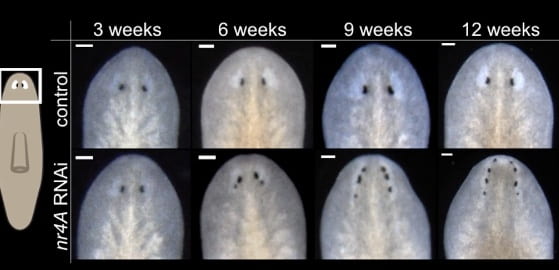
Looking more closely, it turns out that expression domains of PCGs are shifted away from the anterior pole, or head, and expand away from tail tips. This results in the planarian body generating new eyes at shifted regions as a result.
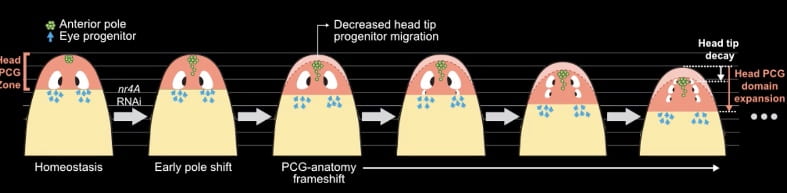
To further verify this, the authors knocked out foxD, which ablates the anterior pole’s identity, and performed RNAi simultaneously on planarians, resulting in a recovery of the wild-type phenotype.
When the authors maintained this process over a longer period of time (131 days), the pattern of eyes never reached a steady state. While new eyes are generated farther from the anterior pole, older eyes get pushed towards the anterior pole and eventually disappear. Thus, the appearance of the nr4A knockdown planarian’s head is constantly in a state of flux. This suggests that eyes are homeostatically maintained in planarians!
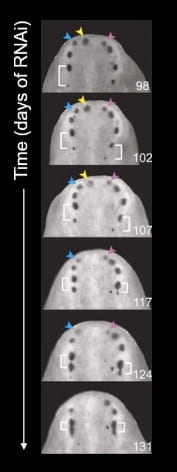
In summary, the authors identified a gene, nr4A, that allows for proper head patterning in planarian flatworms by establishing the domain at which patterning genes can reside in. This contribution is just one of the many that will ultimately lead to us getting a better understanding of how the basic biological process of regeneration works!
You must be logged in to post a comment.