My name is Skyler Sung, a rising third-year Neuroscience major. I look forward to sharing my reflections as a part of the National Institutes of Health Summer Internship Program (NIH SIP). Through this program, students select a lab from one of the many Institutes and Centers (ICs) to work in and are able to participate in talks, courses, and journal clubs hosted by either the Office of Intramural Training or their IC. This summer, I am working remotely with the wonderful Dr. Julie Segre and members of her lab at the National Human Genome Research Institute (NHGRI).
The Segre Lab specializes in clinical microbial genetics, which involves genomic sequencing and analysis to investigate human skin microbiomes and hospital-associated pathogens. Past work in the Segre Lab includes the first skin bacterial survey (characterizing microbe diversity on healthy volunteers) and using genetic sequencing to trace transmission of the 2011 NIH Clinical Center Klebsiella pneumoniae outbreak to guide hospital infection control protocols.
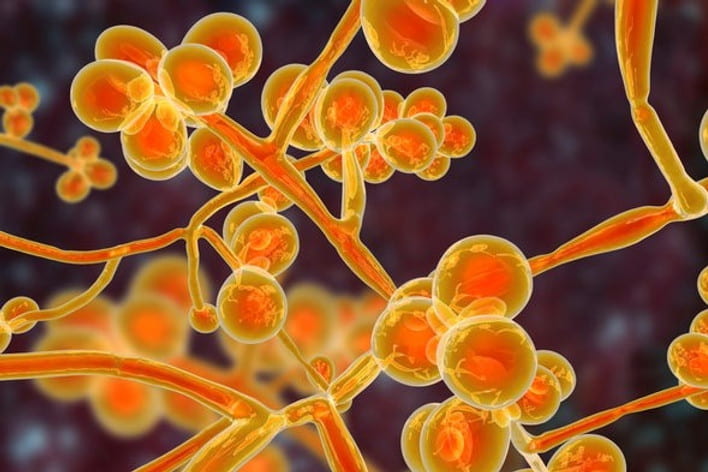
This summer, I will be working on functionally annotating the genome of a Candida auris isolate from a recent outbreak. The emerging fungal pathogen is currently labeled as an urgent global health threat by the CDC for three main reasons: 1) it is often resistant to multiple antifungals 2) basic lab methods can lead to misidentification and thus mistreatment, and 3) it spreads via nosocomial transmission and can cause invasive bloodstream infections (candidiasis) that have a mortality rate in excess of 33%. C. auris, first isolated in Tokyo, has been detected in over 30 countries and is characterized by four clades (populations) that emerged around the same time in four different geographic regions. The yeast can spread quickly throughout healthcare settings via surfaces and on skin, targeting those with weakened immune systems. During the COVID-19 pandemic, there has been an increase in C. auris cases without links to known cases or healthcare abroad, indicating an increase in undetected transmission in the US. There are currently three major classes of antifungals being systematically used against pathogenic Candida: azoles, polyenes, and echinocandins. Although resistance levels vary greatly between clades, each antifungal was met with resistance by at least one clade and a subset of a clade was found to be resistant to all three antifungal classes. Current hypotheses for the recent development of antibiotic resistance include increased use of antifungals in medicine, agriculture, or both. Whole-genome sequencing has helped in characterizing the yeast and its transmission dynamics.
As a remote researcher, I will be using computational tools, such as fungal annotation pipelines over the NIH supercomputing system, to contribute to the field’s understanding of the functional impact of genetic variations in the C. auris genome. By elucidating the genetic code involved in antifungal resistance, skin adhesion, and population structure, researchers and healthcare professionals will better understand the pathogen and how to contain and treat it. I am excited to be one contributing piece to this collaborative global goal.
As a student trained in bench sciences, working on computational biology has involved a great deal of practice using the command line and Python, affording me the opportunity to learn how technology can increase investigative efficiency. I am working remotely in New York, where in my free time I enjoy walking along the Hudson, street photography, and visiting museums.
You must be logged in to post a comment.